New article on arXiv; "Identifying stable communities in Hi-C using multifractal network modularity"
Identifying stable communities in Hi-C using multifractal network modularity
Lucas Hedström, Antón Carcedo Martínez, and Ludvig Lizana,
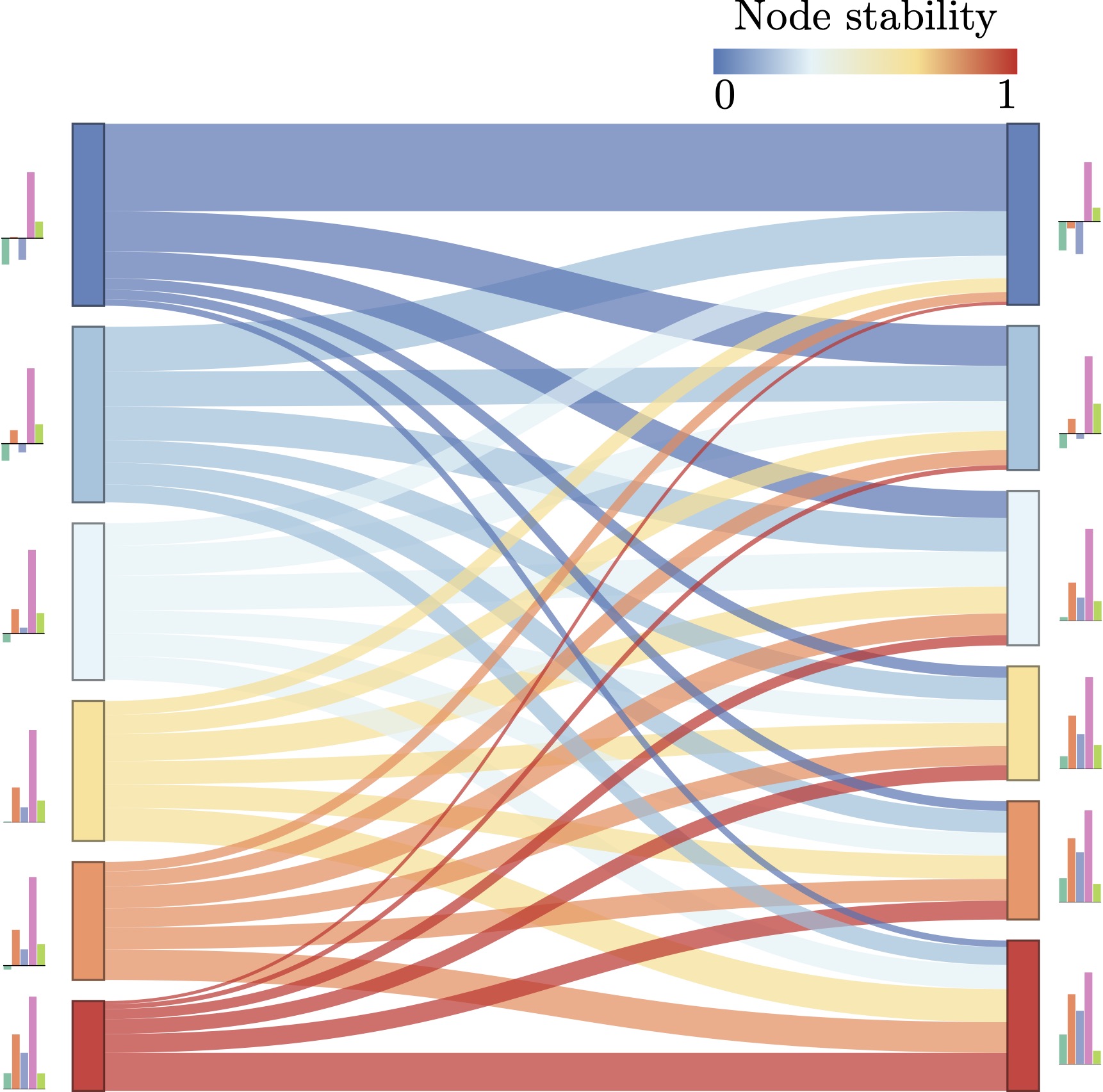
Chromosome capture techniques like Hi-C have expanded our understanding of mammalian genome 3D architecture and how it influences gene activity. To analyze Hi-C data sets, researchers increasingly treat them as DNA-contact networks and use standard community detection techniques to identify mesoscale 3D communities. However, there are considerable challenges in finding significant communities because the Hi-C networks have cross-scale interactions and are almost fully connected. This paper presents a pipeline to distill 3D communities that remain intact under experimental noise. To this end, we bootstrap an ensemble of Hi-C datasets representing noisy data and extract 3D communities that we compare with the unperturbed dataset. Notably, we extract the communities by maximizing local modularity (using the Generalized Louvain method), which considers the multifractal spectrum recently discovered in Hi-C maps. Our pipeline finds that stable communities (under noise) typically have above-average internal contact frequencies and tend to be enriched in active chromatin marks. We also find they fold into more nested cross-scale hierarchies than less stable ones. Apart from presenting how to systematically extract robust communities in Hi-C data, our paper offers new ways to generate null models that take advantage of the network’s multifractal properties. We anticipate this has a broad applicability to several network applications.
@article{hedstrom2024identifying,
title={Identifying stable communities in Hi-C using multifractal network modularity},
author={Lucas Hedstr\"om and Ant\'on Carcedo Mart\'inez and Ludvig Lizana},
journal={arXiv:2405.05425},
year={2024},
}