New plication on PRX Life; "Enhancer-insulator pairing reveals heterogeneous dynamics in long-distant 3D gene regulation"
A general mechanism for enhancer-insulator pairing reveals heterogeneous dynamics in long-distant 3D gene regulation
Lucas Hedström, Ralf Metzler and Ludvig Lizana,
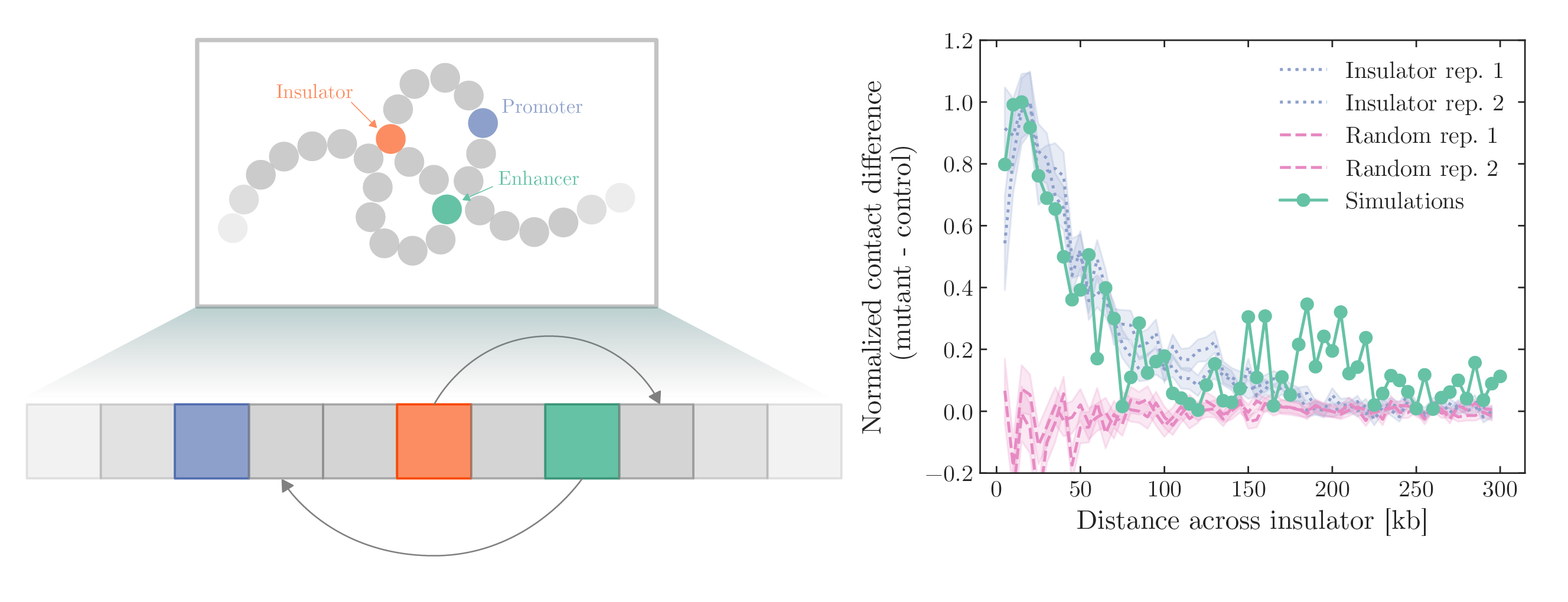
Cells regulate fates and complex body plans using spatiotemporal signaling cascades that al- ter gene expression. Enhancers, short DNA sequences (50-150 base pairs), help coordinate these cascades by attracting regulatory proteins to enhance the transcription of distal genes by bind- ing to promoters. In humans, there are hundreds of thousands of enhancers dispersed across the genome, which poses a challenging coordination task to prevent unintended gene activation. To mit- igate this problem, the genome contains additional DNA elements, insulators, that block enhancer- promoter interactions. However, there is an open problem with how the insulation works, especially as enhancer-insulator pairs may be separated by millions of base pairs. Based on recent empirical data from Hi-C experiments, this paper proposes a new mechanism that challenges the common paradigm that rests on specific insulator-insulator interactions. Instead, this paper introduces a stochastic looping model where enhancers bind weakly to surrounding chromatin. After calibrating the model to experimental data, we use simulations to study the broad distribution of hitting times between an enhancer and a promter when there are blocking insulators. In some cases, there is a large difference between average and most probable hitting times, making it difficult to assign a typical time scale, hinting at highly defocused regulation times. We also map our computational model onto a resetting problem that allows us to derive several analytical results. Besides offering new insights into enhancer-insulator interactions, our paper advances the understanding of gene regulatory networks and causal connections between genome folding and gene activation.
@article{hedstrom2024general,
title={A general mechanism for enhancer-insulator pairing reveals heterogeneous dynamics in long-distant 3D gene regulation},
author={Lucas Hedstr\"om and Ralf Metzler and Ludvig Lizana},
journal={arXiv:2402.09209},
year={2024},
}